Heritability: what it
means for genetic ALS
Genetic ALS / Heritability
What is heritability, and why does it matter to patients with seemingly sporadic ALS?
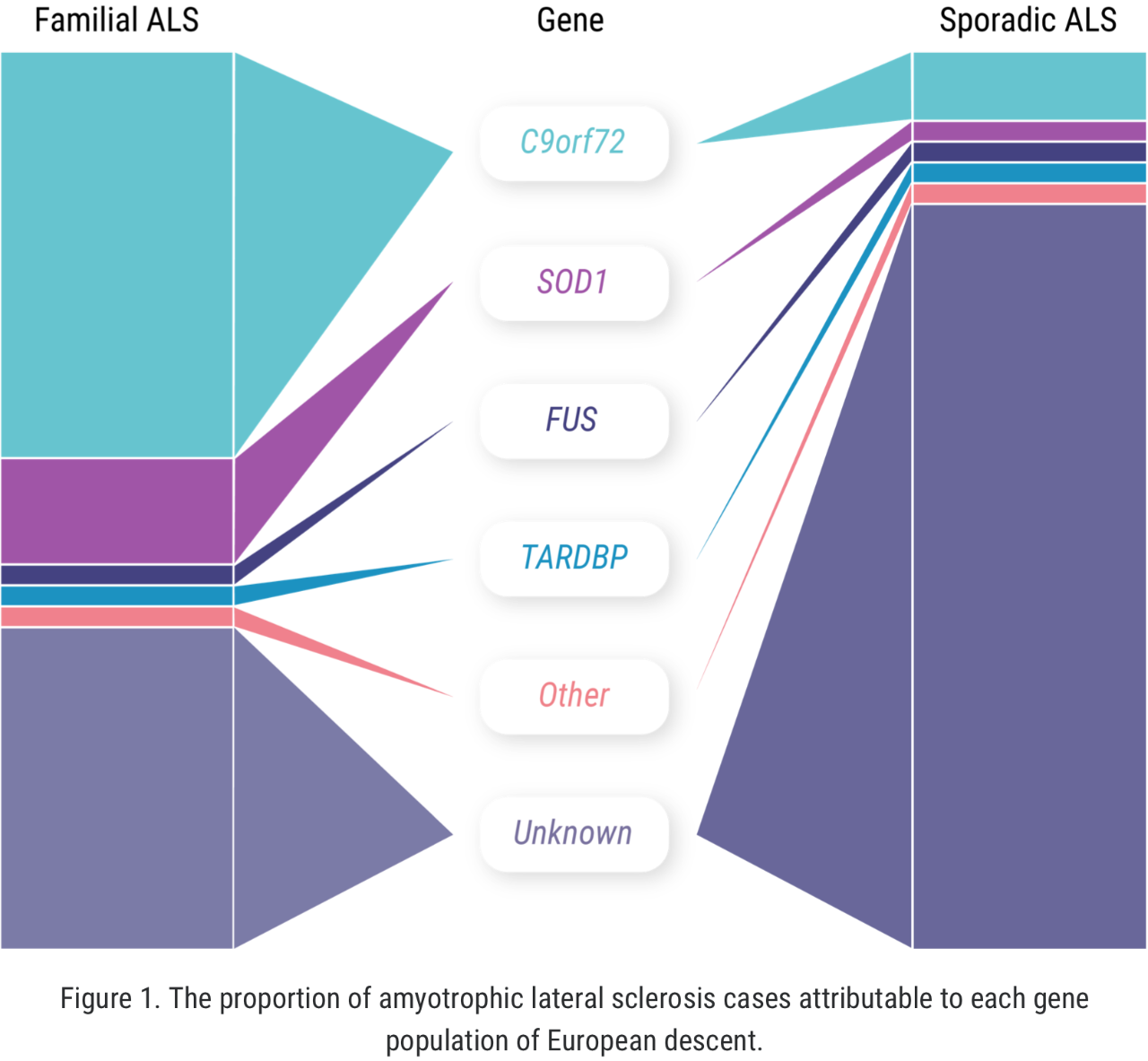
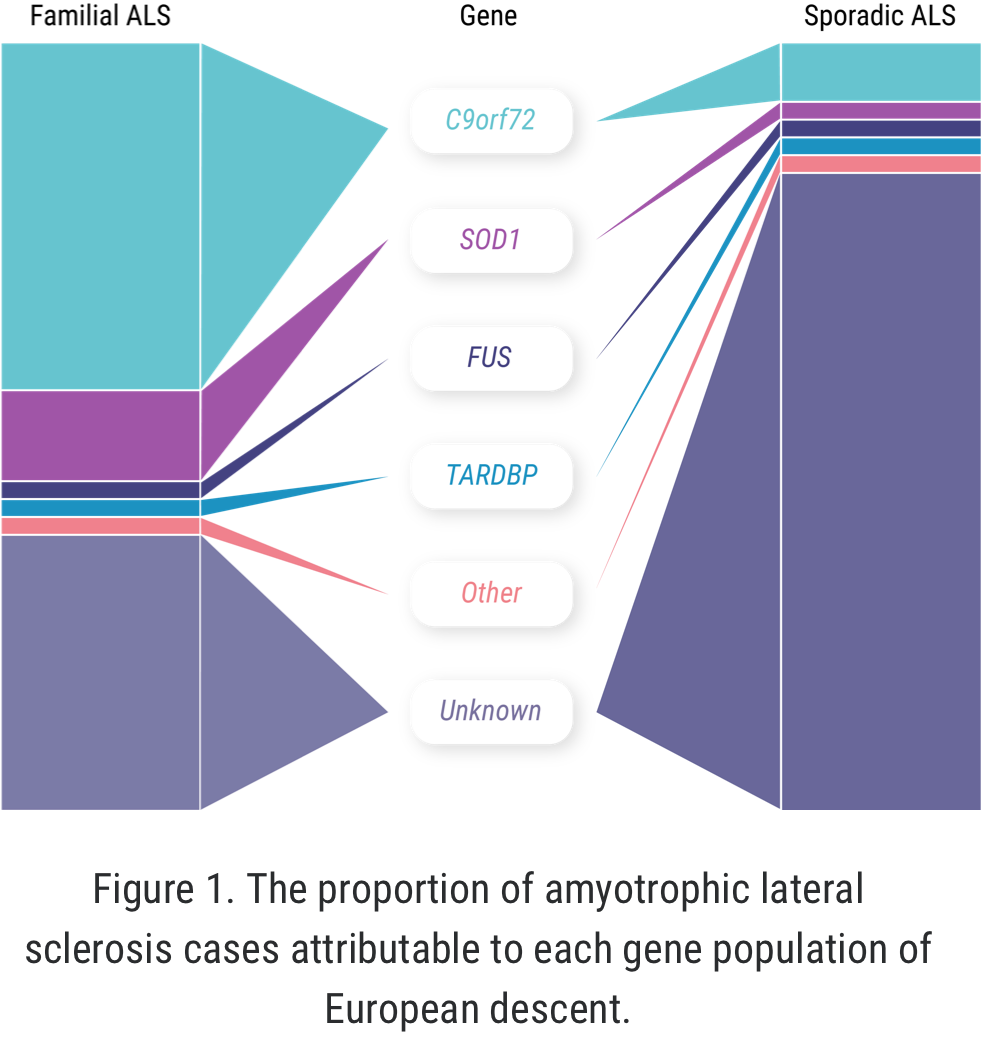
The concept of “familial ALS” is well established: approximately 5% to 10% of all ALS cases occur in patients who have some known family history of the disease or similar neurologic disorders.1,2 But more recently, we have learned that familial ALS is not synonymous with having a genetic driver of the disease. In fact, approximately 10% of seemingly “sporadic” cases of ALS are associated with an identified genetic mutation.3 There may even be more patients with “sporadic ALS” who have a genetic mutation than among those with familial ALS.1-4
Proportion of genetic mutations
This leads to a question from patients who have recently been diagnosed with a genetic form of ALS: “Are my children at risk?”
The answer ties back to the concept of heritability. Research from the Institutes of Neurology and Genetics at Trinity College Dublin has given us new insights into the relationship between genetic ALS and what we call heritability.
What is heritability?
Heritability is the extent to which differences in a given phenotype across a specific population can be accounted for by differences in their genes.5 That population may be as large as a country or ethnic group or as small as a family. Using this concept, the research team identified that genetic factors account for approximately 50% of one’s risk of developing ALS.
Of course, factors outside of a single genetic mutation—such as gender and environmental elements—may also affect whether an individual develops ALS.6
C9orf72: one piece of the genetic ALS puzzle
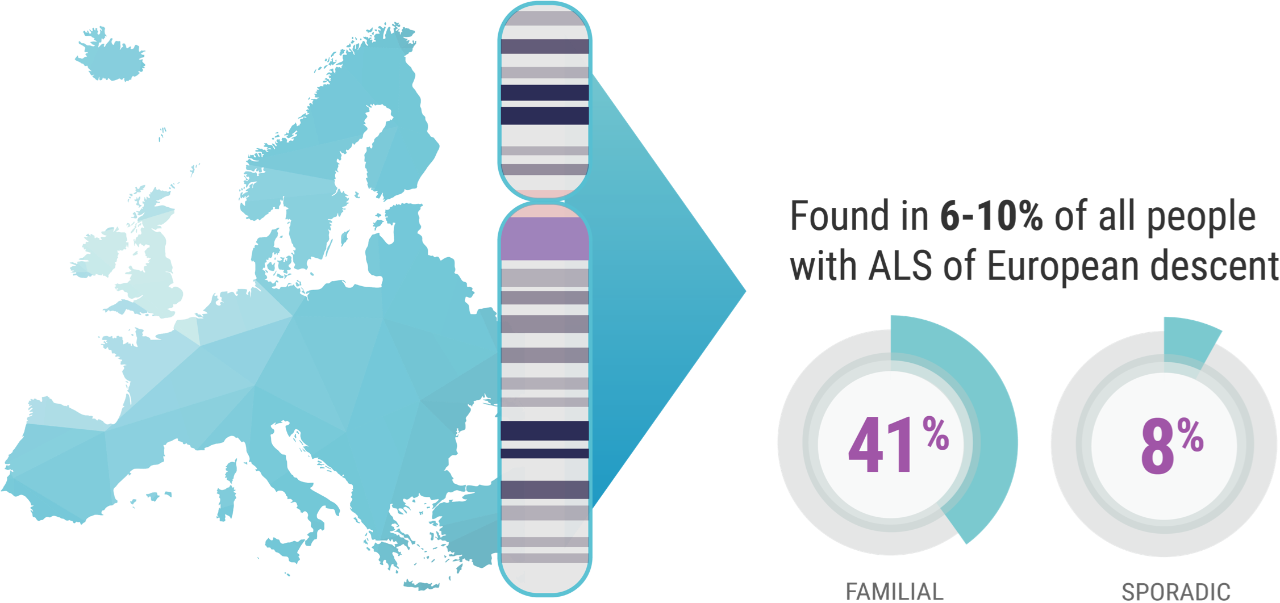
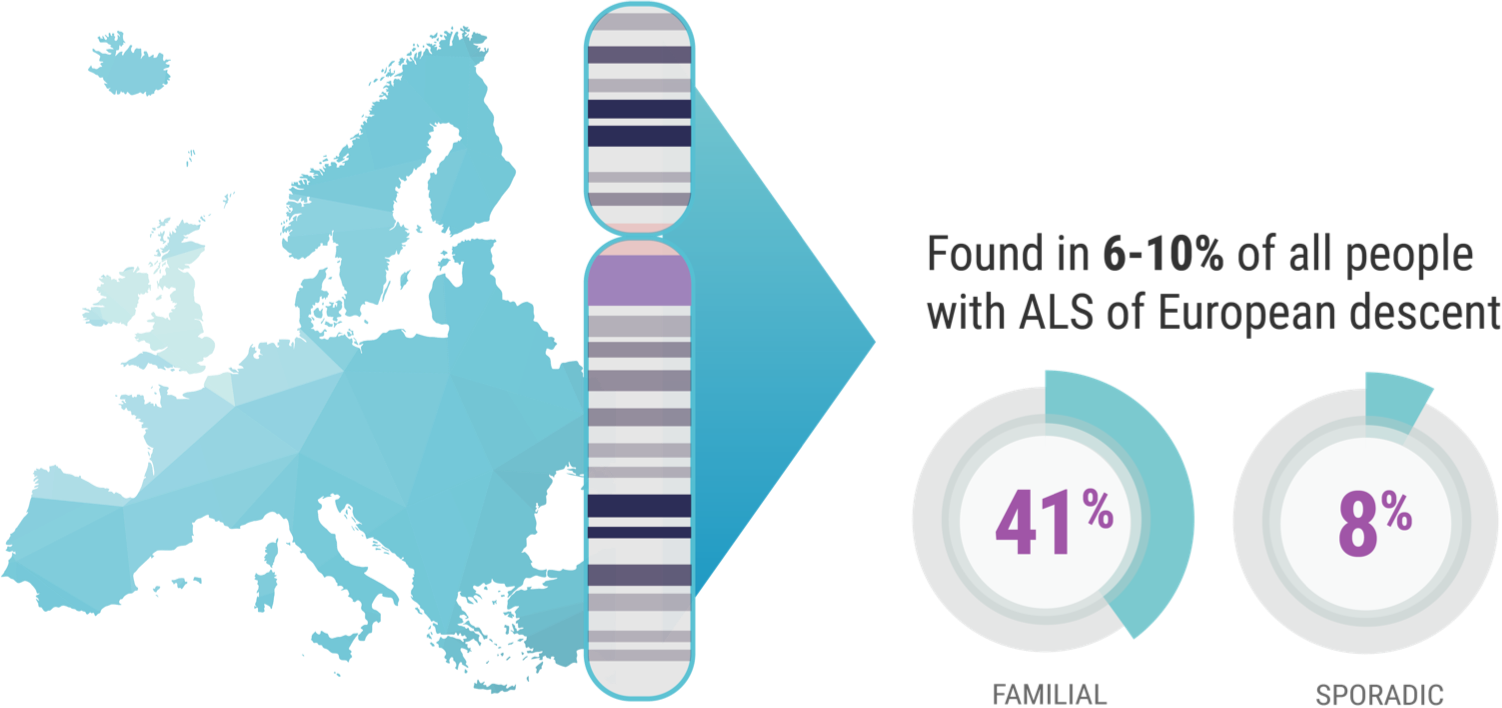
When it comes to C9orf72 and ALS, Professor Orla Hardiman and her team at Trinity College sought to uncover the role of the gene itself, independent of other factors. C9orf72 is the most common gene mutation associated with ALS, affecting 6% to 10% of all patients with ALS of European descent; this includes approximately 41% of all familial ALS and 8% of all sporadic ALS.7,8
Professor Hardiman’s study focused on patients in Ireland who have ALS and the C9orf72 genetic mutation. “And we know,” continued Prof. Hardiman, “for example, 10% of patients in Ireland carry the C9orf72 variant,9 which is just a bit higher than the rest of European-descended populations. So, it's relatively common in Ireland.”
Although SOD1-ALS is the second-most common form of genetic ALS worldwide, no cases have been identified in the Republic of Ireland to date, though cases have been reported in Northern Ireland. Hence, Hardiman and her team focused on C9orf72 as there were no cases of SOD1-ALS in Ireland to investigate.9,10
The study, which looked at parents with C9orf72-ALS and their children, suggested that the presence of the C9orf72 gene mutation was an important determinant of lifetime risk, or the odds of whether a child would develop ALS. This is not to say that having a parent with C9orf72-ALS is a guarantee the child will develop ALS. We still don’t know the true rates of transmission of C9orf72-associated ALS from parents to children.9
According to the team’s research, the lifetime risk of developing ALS for first-degree relatives of individuals with the disease was approximately 1.4% compared with the general population. This is higher than individuals without a first-degree relative with ALS, for whom the rate was approximately 0.3%.9
ALS varies from case to case, based on factors outside of genetic mutations present.11 Hence, the researchers’ second goal was to find out whether the C9orf72 mutation itself contributed to the effect of other factors.
The other pieces of the genetic ALS puzzle
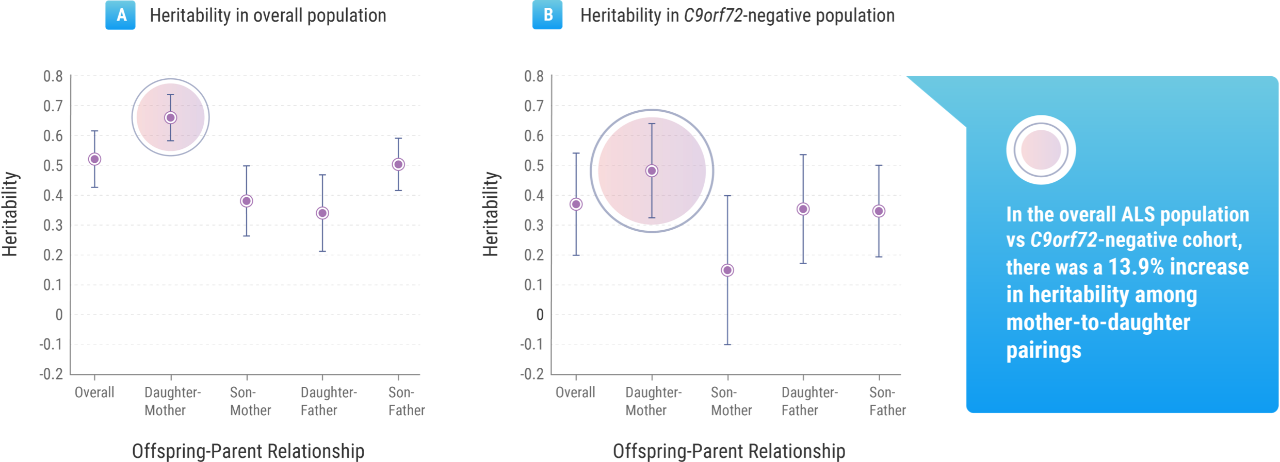
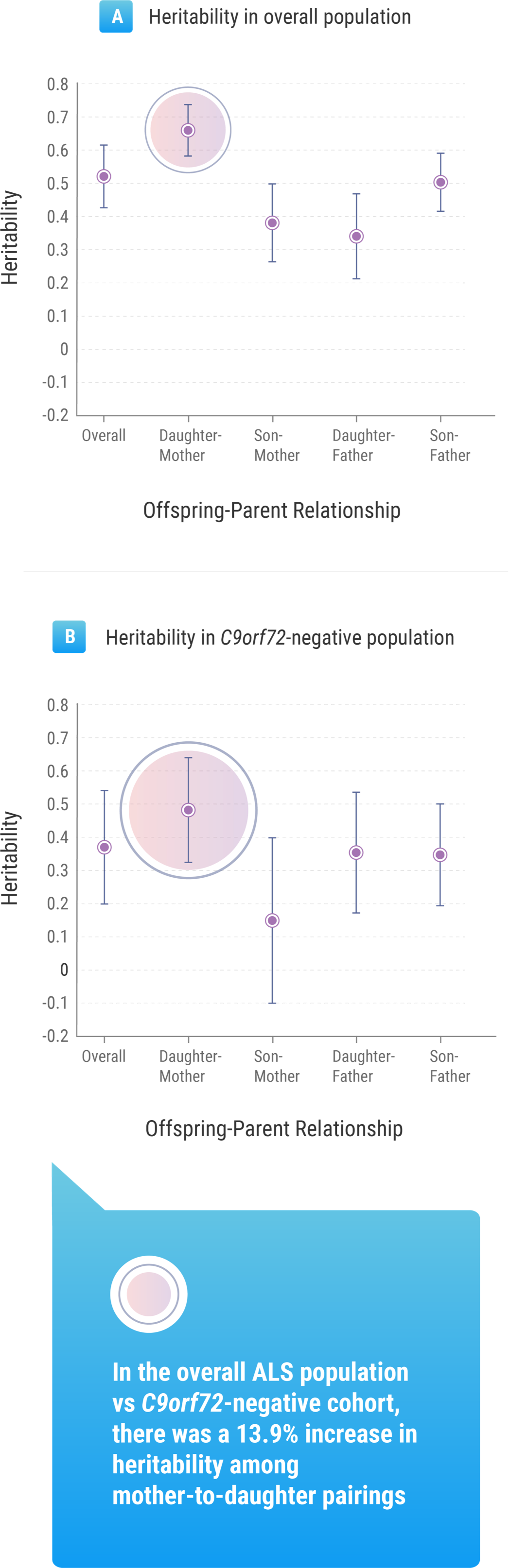
The Trinity College research suggested that gender played a role: the mean lifetime heritability for all patients involved in the study was 52.3%, but this figure rose to 66.2% when considering mother-to-daughter pairings.9 But Prof. Hardiman noted that, beyond these other factors, understanding the genetic impact tied to ALS within a population is important information for researchers such as herself.
“That's a very positive thing in a sense, because if we can understand why that is, or what the patterns are that lead to depressive degeneration, then we're very much on the pathway towards better understanding the disease,” she explained. That understanding relies on identifying more detailed subgroupings of ALS patients to better understand all the factors that play into the complex equation that is the disease’s development, and how it varies from case to case.
Future directions
Further studies will be needed to determine whether the results of this study are applicable to other populations and other mutations such as SOD1-ALS. Prof. Hardiman believes more studies of heritability—and further examination of all the factors that contribute to ALS, along with the interplay between those factors and heritability—are moving ALS research in a positive direction.
“ALS is not unique in the heterogeneity within the disease,” she said. “I think that in a way, our grappling with this level of variability within the disease and ALS is actually coming to the forefront of research in our generation.”
References: 1. Zarei S, Carr K, Reiley L, et al. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol Int. 2015;6:171. 2. Byrne S, Walsh C, Lynch C, et al. Rate of familial amyotrophic lateral sclerosis: a systemic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2011;82:623-627. 3. Volk E, Weishaupt JH, Andersen PM, et al. Current knowledge, and recent insights into the genetic basis of amyotrophic lateral sclerosis. Medgen. 2018;30:252-258. 4. Arthur KC, Calvo A, Price TR, Geiger JT, Chiò A, Traynor BJ. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun. 2016;7:12408. 5. US National Library of Medicine. Genetics. MedlinePlus website. https://medlineplus.gov/genetics/. Accessed July 7, 2021. 6. Vucic S, Westeneng H-J, Al-Chalabi A, Van Den Berg LH, Talman P, Kiernan MC. Amyotrophic lateral sclerosis as a multi-step process: an Australia population study. Amyotrophic Lateral Scler and Frontotemporal Degener. 2019;20(7-8):532-537. doi: 10.1080/21678421.2018.1556697. 7. Ryan M et al. Comparison of the clinical and genetic features of amyotrophic lateral sclerosis across Cuban, Uruguayan and Irish clinic-based populations. J Neurol Neurosurg Psychiatry. 2019 Jun;90(6):659-665. 8. Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323-330. 9. Ryan M, Heverin M, McLaughlin RL, Hardiman O. Lifetime Risk and Heritability of Amyotrophic Lateral Sclerosis. JAMA Neurol. 2019 Jul 22;76(11):1367-74. 10. Roggenbuck J, Quick A, Kolb SJ. Genetic testing and genetic counseling for amyotrophic lateral sclerosis: an update for clinicians. Genet Med. 2017;19(3):267-274. 11. Nguyen HP, Van Broeckhoven C, van der Zee J. ALS genes in the genomic era and their implications for FTD. Trends Genet. 2018;34(6):404-423.
1. Zarei S, Carr K, Reiley L, et al. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol Int. 2015;6:171. 2. Byrne S, Walsh C, Lynch C, et al. Rate of familial amyotrophic lateral sclerosis: a systemic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2011;82:623-627. 3. Volk E, Weishaupt JH, Andersen PM, et al. Current knowledge, and recent insights into the genetic basis of amyotrophic lateral sclerosis. Medgen. 2018;30:252-258. 4. Arthur KC, Calvo A, Price TR, Geiger JT, Chiò A, Traynor BJ. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun. 2016;7:12408. 5. US National Library of Medicine. Genetics. MedlinePlus website. https://medlineplus.gov/genetics/. Accessed July 7, 2021. 6. Vucic S, Westeneng H-J, Al-Chalabi A, Van Den Berg LH, Talman P, Kiernan MC. Amyotrophic lateral sclerosis as a multi-step process: an Australia population study. Amyotrophic Lateral Scler and Frontotemporal Degener. 2019;20(7-8):532-537. doi: 10.1080/21678421.2018.1556697. 7. Ryan M et al. Comparison of the clinical and genetic features of amyotrophic lateral sclerosis across Cuban, Uruguayan and Irish clinic-based populations. J Neurol Neurosurg Psychiatry. 2019 Jun;90(6):659-665. 8. Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323-330. 9. Ryan M, Heverin M, McLaughlin RL, Hardiman O. Lifetime Risk and Heritability of Amyotrophic Lateral Sclerosis. JAMA Neurol. 2019 Jul 22;76(11):1367-74. 10. Roggenbuck J, Quick A, Kolb SJ. Genetic testing and genetic counseling for amyotrophic lateral sclerosis: an update for clinicians. Genet Med. 2017;19(3):267-274. 11. Nguyen HP, Van Broeckhoven C, van der Zee J. ALS genes in the genomic era and their implications for FTD. Trends Genet. 2018;34(6):404-423.